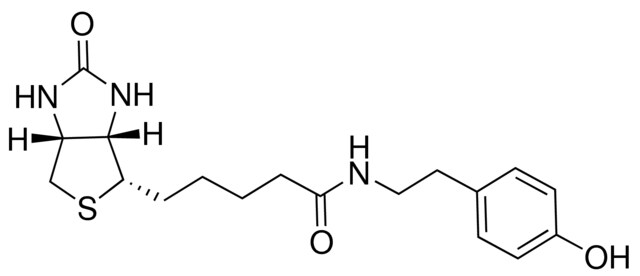
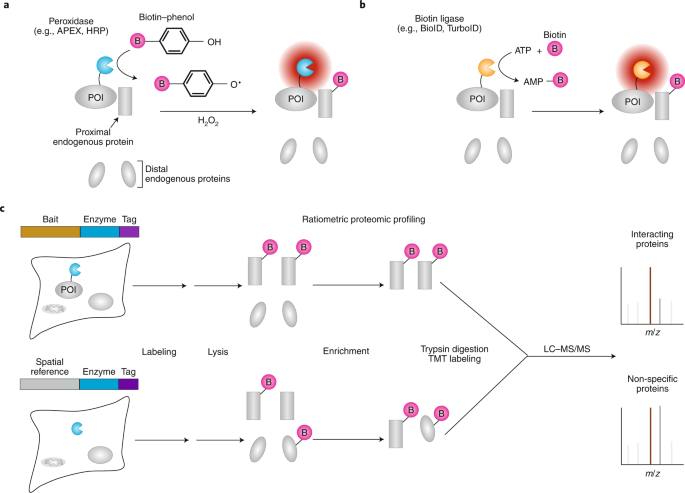
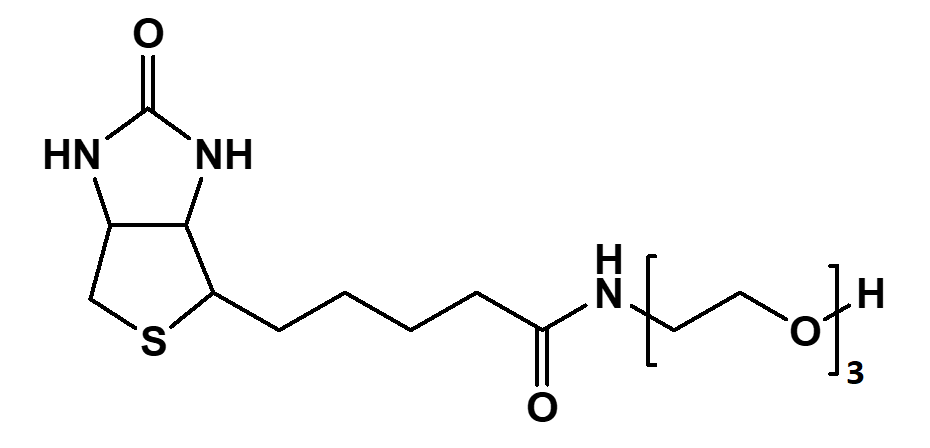
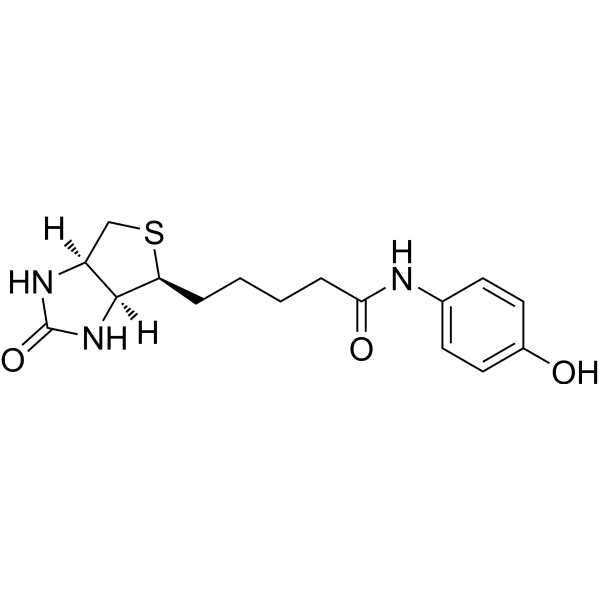
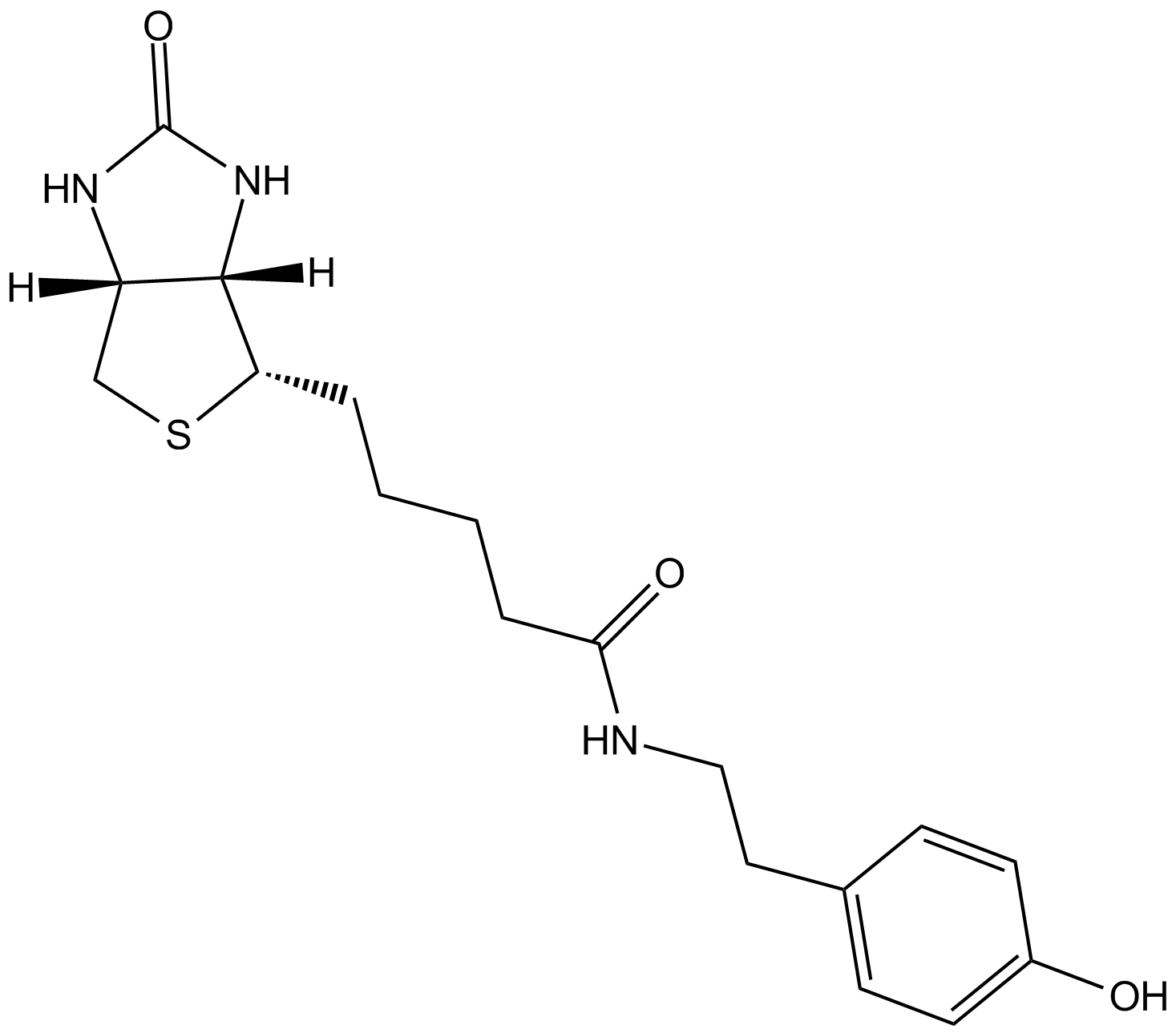
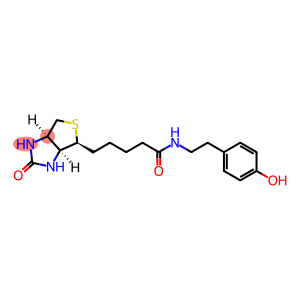
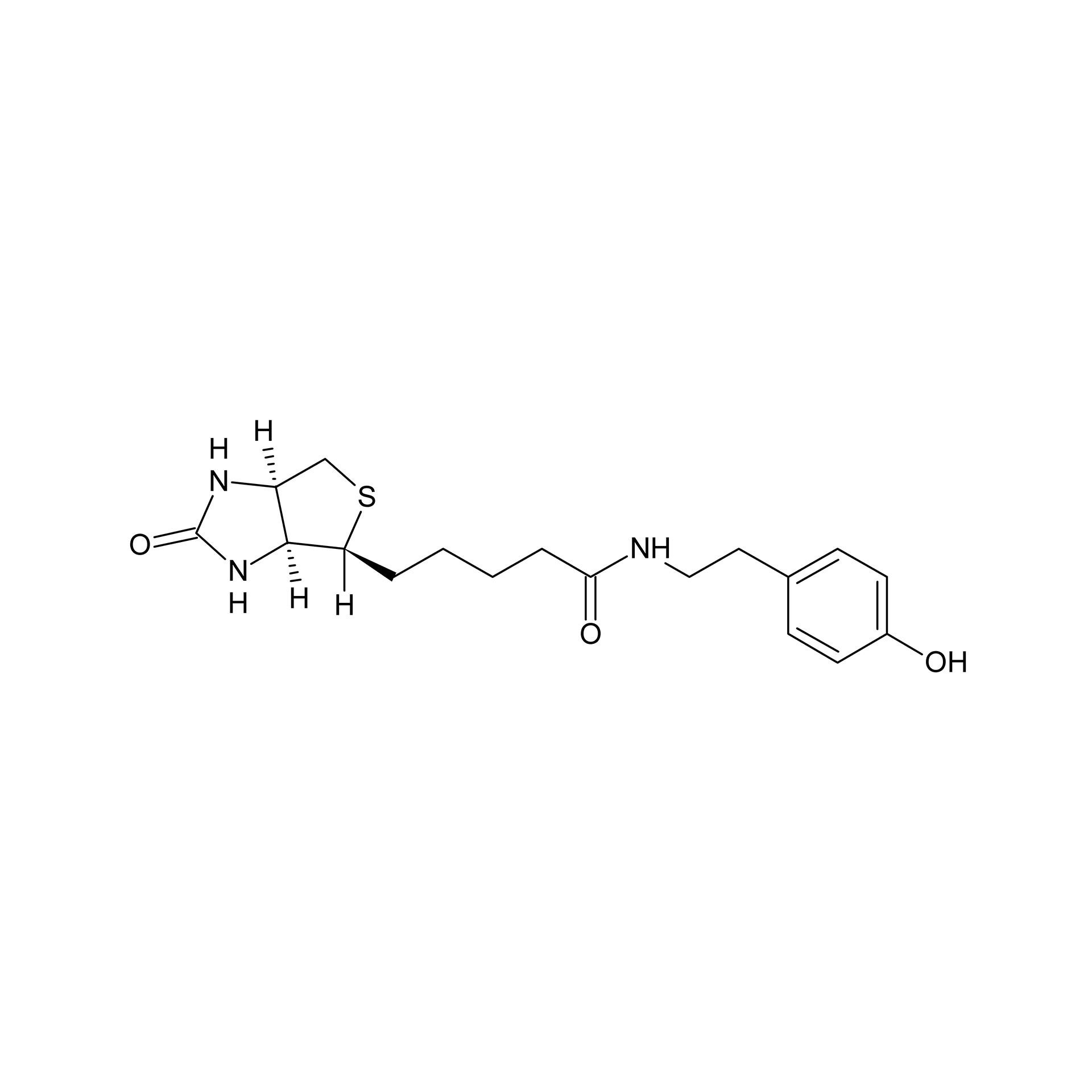
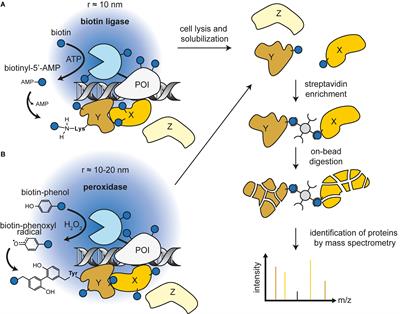
Streptavidin–biotin technology: improvements and innovations in chemical and biological applications | Applied Microbiology and Biotechnology

Uncovering the In Vivo Proxisome Using Proximity‐Tagging Methods - Santin - 2019 - BioEssays - Wiley Online Library

Atlas of Subcellular RNA Localization Revealed by APEX-seq; Proximity RNA labeling by APEX-Seq Reveals the Organization of Translation Initiation Complexes and Repressive RNA Granules - preLights

Proximity Dependent Biotinylation: Key Enzymes and Adaptation to Proteomics Approaches - ScienceDirect
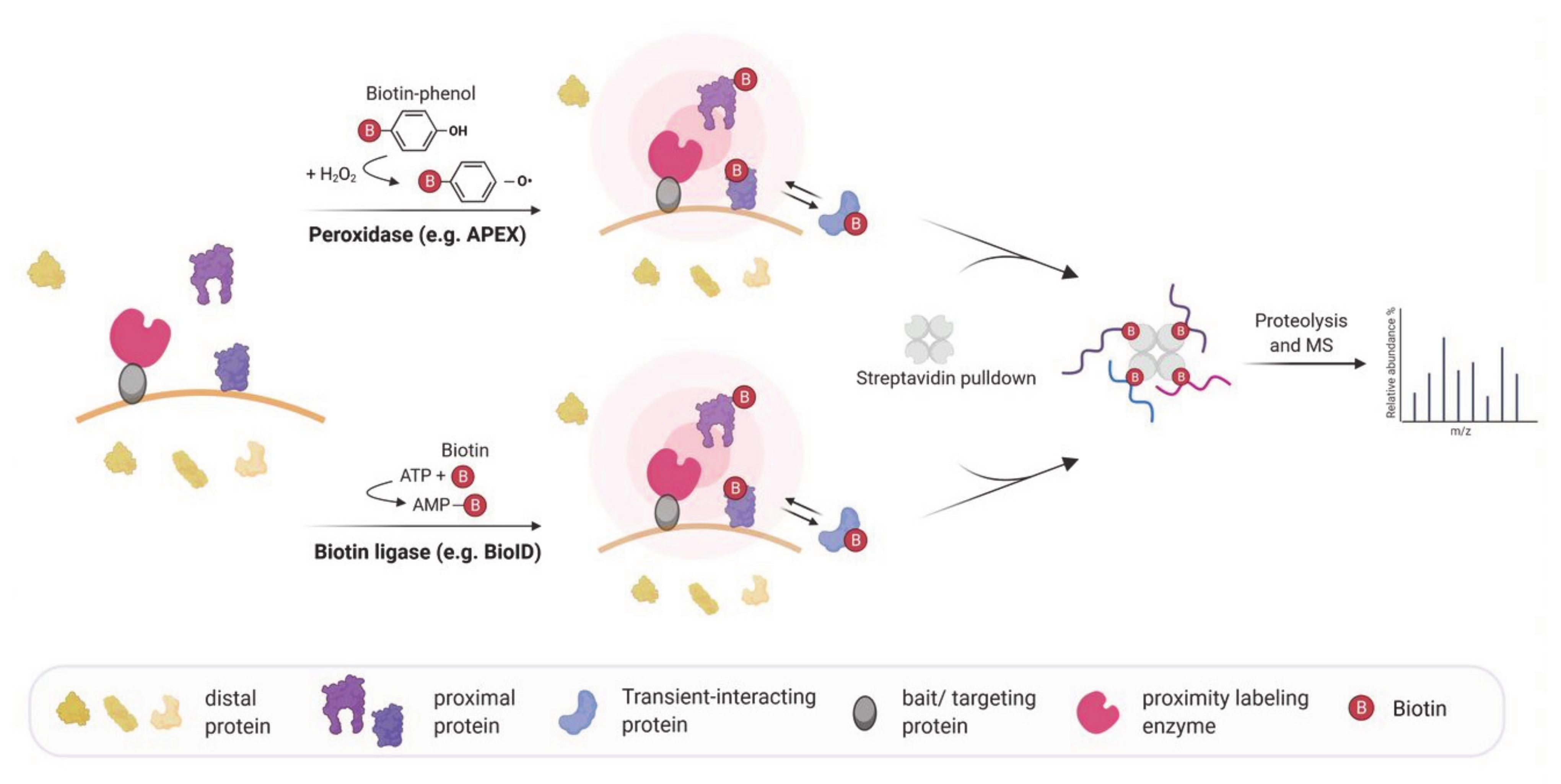
Molecules | Free Full-Text | RNA Proximity Labeling: A New Detection Tool for RNA–Protein Interactions

Schematic representation of the proximity labeling biotinylation of... | Download Scientific Diagram

Fragment ions specific to biotin phenol can be used to enhance spectral... | Download Scientific Diagram

Methods of selective biotinylation. a Selective chemical biotinylation:... | Download Scientific Diagram
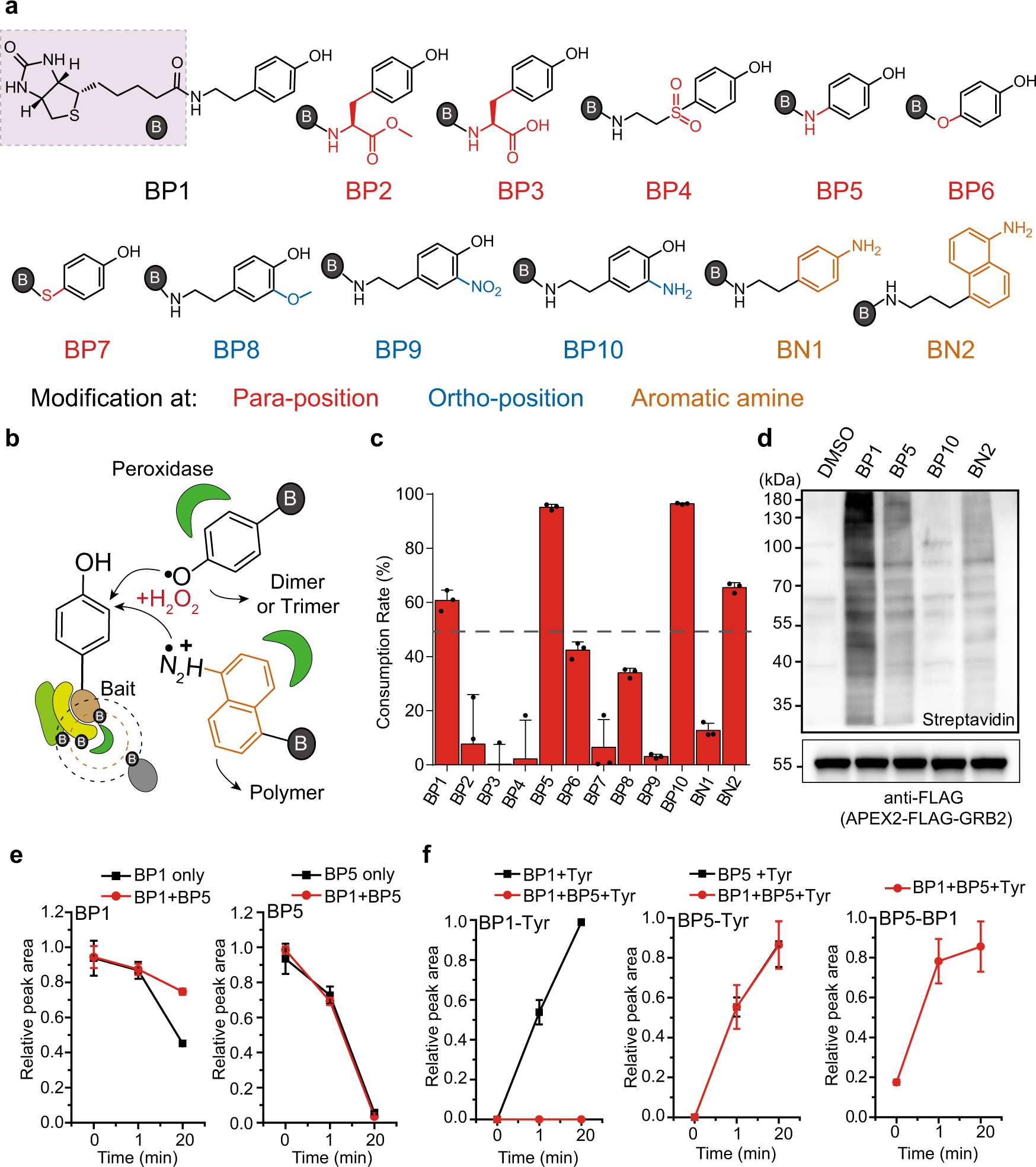
Spatiotemporal profiling of cytosolic signaling complexes in living cells by selective proximity proteomics | Nature Communications

Proximity labeling: an emerging tool for probing in planta molecular interactions. - Abstract - Europe PMC

Expanding APEX2 Substrates for Proximity‐Dependent Labeling of Nucleic Acids and Proteins in Living Cells - Zhou - 2019 - Angewandte Chemie International Edition - Wiley Online Library